Pre-train model
文章来源于:
Paper1:A Systematic Survey of Molecular Pre-trained Models(https://arxiv.org/abs/2210.16484)
Paper2:Mole-BERT: Rethinking Pre-training Graph Neural Networks for Molecules(Mole-BERT: Rethinking Pre-training Graph Neural Networks for Molecules | OpenReview)
文章汇总表:
| Model | Architecture | Inputs | Pre-training Database | 下游任务数据 | 是否有代码 | |
| SEQUENCE | SMILES Transformer | Transformer | SMILES | ChEMBL (861k) | molecular property prediction.(10 datasets) • Physical chemistry: ESOL(1128), FreeSolv(643), and Lipophilicity(4200) • Biophysics: MUV(93127), HIV(41913), and BACE(1522) • Physiology: BBBP(2053), Tox21(8014), SIDER(1427), and ClinTox(1491) | Link |
| ChemBERTa | Transformer | SMILES/SELFIES | PubChem (77M) | masked prediction tasks, fine-tuning the model on the Tox21 SR-p53(7831) dataset. | Link | |
| SMILES-BERT | Transformer | SMILES | ZINC15(∼18.6M) | Molecular Property Prediction Small scale dataset (LogP 10850) to large-scale datasets (PM2 324232 and PCBA 686978) | Link | |
| Molformer | Transformer | SMILES | ZINC15(1B) + PubChem(111M) | Classification Tasks: BBBP,ClinTox,HIV,BACE,SIDER Regression Tasks:QM9, QM8, ESOL, FreeSolv, and Lipophilicity | - | |
| GRAPH & GEOMETRY | Hu et al. | 5-layer GIN | Graph | ZINC15(2M) + ChEMBL(456K) | molecular property prediction: BBBP(2039),Tox21(7831),ToxCast(8575),SIDER(1427),ClinTox(1478),MUV(93087),HIV(41127),BACE(1513) | Link |
| GraphCL | 5-layer GIN | Graph | ZINC15(2M) + ChEMBL(456K) | NCI1 PROTEINS DD COLLAB RDT-B RDT-M5K GITHUB MNIST CIFAR10 | Link | |
| JOAO | 5-layer GIN | Graph | ZINC15(2M) + ChEMBL(456K) | BBBP Tox21 ToxCast SIDER ClinTox MUV HIV BACE PPI | Link | |
| AD-GCL | 5-layer GIN | Graph | ZINC15(2M) + ChEMBL(456K) | chemical molecules property prediction: BBBP,Tox21,SIDER,ClinTox,BACE,HIV,MUV,ToxCast | Link | |
| GraphLog | 5-layer GIN | Graph | ZINC15(2M) + ChEMBL(456K) | molecular property prediction: BBBP,Tox21,ToxCast,SIDER,ClinTox,MUV,HIV,BACE | Link | |
| MGSSL | 5-layer GIN | Graph | ZINC15 (250K) | molecular property prediction: muv,clintox,sider,hiv,tox21,bace,toxcast,bbbp | Link | |
| MPG | MolGNet | Graph | ZINC + ChEMBL (11M) | molecular properties prediction: Tox21(7831),ToxCast(8575),SIDER(1427),ClinTox(1478),BACE(1513),BBBP(2039),FreeSolv(642),ESOL(1128),Lipo(4200) | Link | |
| LP-Info | 5-layer GIN | Graph | ZINC15(2M) + ChEMBL(456K) | Eight fine-tuning/evaluation datasets are taken from MoleculeNet(BBBP,Tox21,ToxCast,SIDER,ClinTox,MUV,HIV,BACE) | Link | |
| SimGRACE | 5-layer GIN | Graph | ZINC15(2M) + PPI(306K) | BBBP(2039), ToxCast(8576) and SIDER(1427). | Link | |
| GraphMAE | 5-layer GIN | Graph | ZINC15(2M) + ChEMBL(456K) | Finetuned in 8 classification benchmark datasets contained in MoleculeNet | Link | |
| GROVER | GTransformer | Graph | ZINC + ChEMBL (10M) | Classification:BBBP(2039),SIDER(1427),ClinTox(1478),BACE(1513),Tox21(7831),ToxCast(8575) Regression:FreeSolv(642),ESOL(1128),Lipo(4200),QM7(6830),QM8(21786) | Link | |
| MolCLR | GCN + GIN | Graph | PubChem (10M) | Seven classification :BBBP(2039),Tox21(7831),ClinTox(1478),HIV(41127),BACE(1513),SIDER(1427),MUV(93087) Six regression :FreeSolv(642),ESOL(1128),Lipo(4200),QM7(6830),QM8(21786),QM9(130829) | Link | |
| Graphormer | Graphormer | Graph | PCQM4M-LSC (∼3.8M) | Binary classification:OGBG-MolPCBA(437,929),OGBG-MolHIV(41,127), Regression:ZINC (sub-set)(12000) | Link | |
| Denoising | GNS | Geometry | PCQM4Mv2(∼3.4 M) | Molecular property prediction:QM9(130K) | Link | |
| InfoGraph | GIN{4,8,12} | Graph | Graph classification(MUTAG, PTC, REDDIT-BINARY, REDDIT-MULTI-5K, IMDB-BINARY,IMDB-MULTI) + semi-supervised molecular property prediction tasks(QM9(130K)) | Link | ||
| EdgePred | Graph | Reddit, WoS, PPI(50k edges) | Link | |||
| GPT-GNN | GPT-GNN | Graph | Open Academic Graph (OAG) of 179 million nodes & 2 billion edges and Amazon recommendation data of 113 million nodes. | OAG(Paper–Field, Paper–Venue, Author ND) + Amazon( Fashion, Beauty, Luxury) 10% of labeled data is used for fine-tuning | Link | |
| ContextPred | 5-layer GIN | Graph | ZINC15(2M) + ChEMBL(456K) | molecular prediction: BBBP(2039),Tox21(7831),ToxCast(8575),SIDER(1427),ClinTox(1478),MUV( 93087),HIV(41127),BACE(1513) | Link | |
| MULTIMODAL/KNOWLEDGE-ENRICHED | DMP | DeeperGCN + Transformer | Graph + SMILES | PubChem (110M) | Molecular property prediction: BBBP(2039),Tox21(7831),ClinTox(1478),HIV(41127),BACE(1513),SIDER(1478) | Link |
| GraphMVP | 5-layer GIN + SchNet | Graph + Geometry | GEOM (50k) | molecular property prediction tasks: BBBP Tox21 ToxCast Sider ClinTox MUV HIV Bace Avg | Link | |
| 3D Infomax | PNA | Graph + Geometry | QM9(50K) + GEOM(140K) + QMugs(620K) | molecular property prediction tasks: Fine tuned on molecules from QM9(50k) from GEOM-Drugs(140k) | Link | |
| KCL | GCN + KMPNN | Graph + KG | ZINC15 (250K) | We use 8 benchmark datasets from the MoleculeNet: Classification (ROC-AUC):BBBP(2039),Tox21(7831),ToxCast(8575),SIDER(1427),ClinTox(1478),BACE(1513) Regression (RMSE):ESOL(1128),FreeSolv(642) | Link | |
| KV-PLM | Transformer | SMILES + Text | PubChem(150M)+S2orc | cross retrieval between substances and property descriptions.(PCdes)(15k SMILES-description pairs in PCdes) | Link | |
| MOCO | Transformer + GIN + SchNet | SMILES + FP + Graph + Geometry | GEOM(50k) | Physical chemistry: ESOL(1128),Lipophilicity(4200),CEP(29978) Biophysics: HIV(41127),MUP(93087),BACE(1513),Malaria(9999) Physiology: BBBP(2039),Tox21(7831),ToxCast(8576),ClinTox(1477),SIDER(1427) Quantum properties: QM9(130831) | - | |
| UniMol | Transformer | Geometry + Protein Pockets | ZINC/ChemBL + PDB | Molecular property prediction(15 datasets):BBBP,BACE,ClinTox,Tox21,ToxCast,SIDER,HIV,PCBA,MUV,ESOL,FreeSolv,lipo,QM7, QM8, QM9 Molecular corformation generation:GEOM Pocket property prediction:NRDLD,Our created benchmark dataset The dataset contains 164,586 candidate pockets Protein-ligand binding pose prediction:PDBbind General set v.2020,CASF-2016 | Link | |
| MolT5 | Transformer | SMILES + Text | C4 + ZINC-15 (100 M) | Molecule Generation,Molecule Captioning(ChEBI-20(33K)) | Link | |
| MICER | CNNs + LSTM | SMILES + Image | ZINC20 | Link | ||
| MM-Deacon | Transformer | SMILES + IUPAC | PubChem | Molecular property prediction: Classification tasks:BBBP(2039),ClinTox(1478),HIV(41127),SIDER(1427), Regression tasks:ESOL(1128),FreeSolv(642),Lipophilicity(4200) | - | |
| PanGu Drug Model(没有代码) | Transformer + TransformerConv | Graph + SELFIES | ZINC20+ DrugSpaceX+UniChem(∼1.7 B) | Link | ||
| ChemRL-GEM(用paddle写的) | GeoGNN | Graph + Geometry | ZINC15 (20M) | Regression:ESOL(1128) FreeSolv(642) Lipo(4200) QM7(6830) QM8(21786) QM9(133885) Classifcation:BACE(1513) BBBP(2039) ClinTox(1478) SIDER(1427) Tox21(7831) ToxCast(8575) HIV(41127) MUV(93087) PCBA(437929) | Link |
模型介绍:
1、SMILES Transformer
introduction
SMILES Transformer extracts molecular fingerprints from string representations of chemical molecules.
The transformer learns latent representation that is useful for various downstream tasks through autoencoding task.
Dataset
Canonical SMILES of 1.7 million molecules that have no more than 100 characters from Chembl24 dataset were used.
These canonical SMILES were transformed randomly every epoch with SMILES-enumeration by E. J. Bjerrum.
备注:关于SMILES enumeration(SMILES enumeration, vectorization and batch generation):
SMILES enumeration is the process of writing out all possible SMILES forms of a molecule. It's a useful technique for data augmentation before sequence based modeling of molecules. You can read more about the background in this blog post or this preprint on arxiv.org
pretrain-gnns
【只要它的数据处理部分】:这里直接把下载的数据的toxcast文件夹复制到MoLClR的data下面就行了
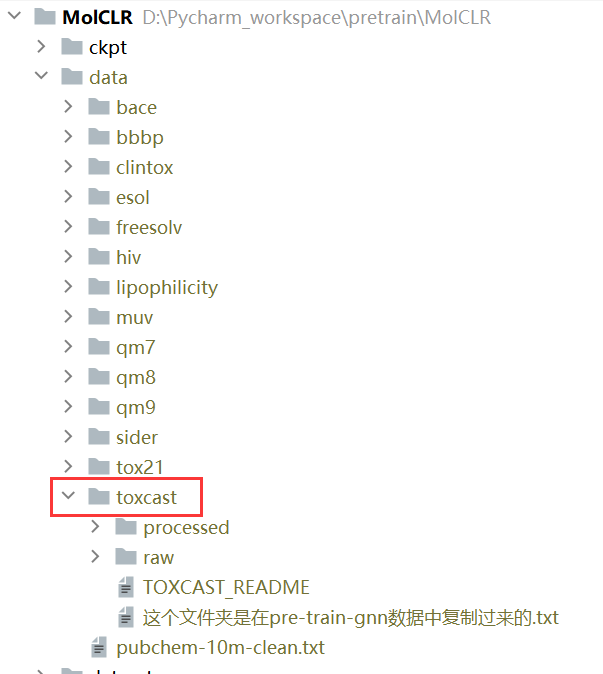
1、MoLClR:
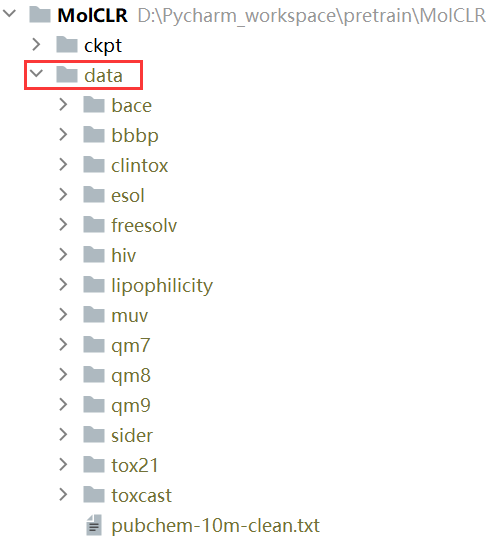
数据准备:
You can download the pre-training data and benchmarks used in the paper here and extract the zip file under ./data folder. The data for pre-training can be found in pubchem-10m-clean.txt.
环境安装:
# create a new environment
$ conda create --name molclr python=3.7
$ conda activate molclr# install requirements
$ pip install torch==1.7.1+cu110 torchvision==0.8.2+cu110 -f https://download.pytorch.org/whl/torch_stable.html
$ pip install torch-geometric==1.6.3 torch-sparse==0.6.9 torch-scatter==2.0.6 -f https://pytorch-geometric.com/whl/torch-1.7.0+cu110.html$ pip install torch-geometric==1.5.0
$ pip install PyYAML
$ conda install -c conda-forge rdkit=2020.09.1.0
$ conda install -c conda-forge tensorboard# clone the source code of MolCLR
$ git clone https://github.com/yuyangw/MolCLR.git
$ cd MolCLR
downstream:
To fine-tune the MolCLR pre-trained model on downstream molecular benchmarks, where the configurations and detailed explaination for each variable can be found in config_finetune.yaml
要注意: config_finetune.yaml中的 task_name: BBBP 要大写而不要bbbp
cd MolCLR
python finetune.py
2、MGSSL
GitHub - zaixizhang/MGSSL: Official implementation of NeurIPS'21 paper"Motif-based Graph Self-Supervised Learning for Molecular Property Prediction"
修改地方为:
1.模型修改
model.py中将386行进行修改,修改为如下(也就说直接跳过pretrain的过程,然后直接finetune就可以了,否则会出现“No such file or directory: '../motif_based_pretrain/saved_model/motif_pretrain.pth'”这种错误):
def from_pretrained(self, model_file):# #self.gnn = GNN(self.num_layer, self.emb_dim, JK = self.JK, drop_ratio = self.drop_ratio)# self.gnn.load_state_dict(torch.load(model_file))pass2.数据集复制
另外我将MPG模型中的“data/downstream”的文件夹复制过来,然后修改为dataset/....

3.finetune数据修改
finetune.py中的109行替换 default即可:
parser.add_argument('--dataset', type=str, default = 'bbbp', help='root directory of dataset. For now, only classification.')运行:
cd ../MGSSL
python finetune/finetune.py
3、MPG
https://github.com/pyli0628/MPG
报错 1:
Number of parameter: 53.32M
====epoch 1
Iteration: 0%| | 0/429 [00:00 Traceback (most recent call last):
File "property/finetune.py", line 297, in
main()
File "property/finetune.py", line 254, in main
train(args, model, device, train_loader, optimizer,criterion)
File "property/finetune.py", line 45, in train
pred = model(batch)
File "/home/mapengsen/anaconda3/envs/grover/lib/python3.6/site-packages/torch/nn/modules/module.py", line 889, in _call_impl
result = self.forward(*input, **kwargs)
File "/mnt/d/Pycharm_workspace/pretrain/MPG/property/model.py", line 292, in forward
node_representation = self.gnn(x, edge_index, edge_attr,node_seg,edge_seg)
File "/home/mapengsen/anaconda3/envs/grover/lib/python3.6/site-packages/torch/nn/modules/module.py", line 889, in _call_impl
result = self.forward(*input, **kwargs)
File "/mnt/d/Pycharm_workspace/pretrain/MPG/property/model.py", line 223, in forward
x = gnn(x,edge_index,edge_attr)
File "/home/mapengsen/anaconda3/envs/grover/lib/python3.6/site-packages/torch/nn/modules/module.py", line 889, in _call_impl
result = self.forward(*input, **kwargs)
File "/mnt/d/Pycharm_workspace/pretrain/MPG/property/model.py", line 175, in forward
attention_output = self.attention.forward(x, edge_index, edge_attr)
File "/mnt/d/Pycharm_workspace/pretrain/MPG/property/model.py", line 147, in forward
return self.propagate(edge_index, size=size, x=x, pseudo=pseudo)
File "/home/mapengsen/anaconda3/envs/grover/lib/python3.6/site-packages/torch_geometric/nn/conv/message_passing.py", line 317, in propagate
out = self.message(**msg_kwargs)
File "/mnt/d/Pycharm_workspace/pretrain/MPG/property/model.py", line 155, in message
alpha = softmax(alpha, edge_index_i, size_i)
RuntimeError: softmax() Expected a value of type 'Optional[Tensor]' for argument 'ptr' but instead found type 'int'.
Position: 2
Value: 362
Declaration: softmax(Tensor src, Tensor? index=None, Tensor? ptr=None, int? num_nodes=None, int dim=0) -> (Tensor)
Cast error details: Unable to cast Python instance to C++ type (compile in debug mode for details)
(grover) mapengsen@DESKTOP-RMA9452:/mnt/d/Pycharm_workspace/pretrain/MPG$ pip list
Package Version
--------------------- -----------
ase 3.22.1
certifi 2021.5.30
首次运行出现了上述的问题,然后我不知道到底是哪里的问题,然后我不知道怎么回事,一直以为是softmax的问题(这个是根据readme走下来的,我们认为readme一般是没有问题的,出现错误多半是因为安装的版本有问题),所以根据出现的所有的错误,我将错误定位在了倒数第二个错误,其中site-packages/下面包含了torch_geometric,所以这里我们认为是torch_geometric出现了错误,然后conda list 发现torch-geometric版本装成了2.0以上的,应该装为readme一样的版本1.5才可以,
解决: pip install torch-geometric==1.5.0
报错2:
Traceback (most recent call last):
File "property/finetune.py", line 297, in
main()
File "property/finetune.py", line 204, in main
dataset = MoleculeDataset(args.data_dir + args.dataset, dataset=args.dataset,transform=transform
File "/mnt/d/Pycharm_workspace/pretrain/MPG/property/loader.py", line 501, in __init__
self.data, self.slices = torch.load(self.processed_paths[0])
File "/home/mapengsen/anaconda3/envs/grover/lib/python3.6/site-packages/torch/serialization.py", line 592, in load
return _load(opened_zipfile, map_location, pickle_module, **pickle_load_args)
File "/home/mapengsen/anaconda3/envs/grover/lib/python3.6/site-packages/torch/serialization.py", line 851, in _load
result = unpickler.load()
ModuleNotFoundError: No module named 'torch_geometric.data.storage'
(grover) mapengsen@DESKTOP-RMA9452:/mnt/d/Pycharm_workspace/pretrai
ModuleNotFoundError: No module named 'torch_geometric.data.storage'
解决:只需要把原来processed文件全部删除就可以了:为什么这么做?
因为我们发现最后报出的错误是No module named 'torch_geometric.data.storage',其中storage一般是用的存储相关的,基于上述错误,我们认为应该是上次运行torch_geometric出现了错误,将错误的文件保存在了本地,所以我们直接删除上次运行产生的文件,就可以运行了
运行:
cd MPG
python property/finetune.py --data_dir data/downstream/ --dataset toxcast --lr 0.0001
4、GraphMVP
首先环境安装:
pip install torch==1.9.1+cu111 torchvision==0.10.1+cu111 torchaudio==0.9.1 -f https://download.pytorch.org/whl/torch_stable.htmlconda create -n GraphMVP python=3.7
conda activate GraphMVPconda install -y -c rdkit rdkit
conda install -y numpy networkx scikit-learn
pip install ase
pip install git+https://github.com/bp-kelley/descriptastorus
pip install ogb
pip install torch_cluster==1.5.9-cp37-cp37m-linux_x86_64.whl
pip install torch_scatter==2.0.9-cp37-cp37m-linux_x86_64.whl
pip install torch_sparse==0.6.12-cp37-cp37m-linux_x86_64.whl
pip install torch-geometric==1.7.2
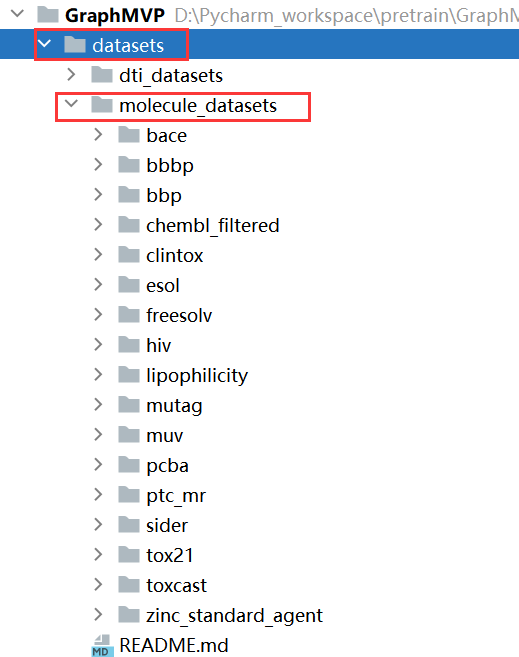
数据下载:
wget http://snap.stanford.edu/gnn-pretrain/data/chem_dataset.zip
unzip chem_dataset.zip
将下载的数据文件夹名字改为molecule_datasets,然后放在dataset文件夹下面
运行:
cd ../GraphMVP/src_classification/
python molecule_finetune.py
5、grover
code:GitHub - tencent-ailab/grover: This is a Pytorch implementation of the paper: Self-Supervised Graph Transformer on Large-Scale Molecular Data
将basemodel权重下载:https://ai.tencent.com/ailab/ml/ml-data/grover-models/pretrain/grover_base.tar.gz
并放在“model/tryout/grover_base.pt”中
command:
python main.py finetune --data_path exampledata/finetune/bbbp.csv --save_dir model/finetune/bbbp/ --checkpoint_path model/tryout/grover_base.pt --dataset_type classification --split_type scaffold_balanced --ensemble_size 1 --num_folds 3 --no_features_scaling --ffn_hidden_size 200 --batch_size 32 --epochs 10 --init_lr 0.00015
或者直接finetune:
python main.py finetune --data_path exampledata/finetune/bbbp.csv --save_dir model/finetune/bbbp/ --hidden_size 3 --dataset_type classification --split_type scaffold_balanced --ensemble_size 1 --num_folds 3 --no_features_scaling --ffn_hidden_size 200 --batch_size 32 --epochs 10 --init_lr 0.00015
